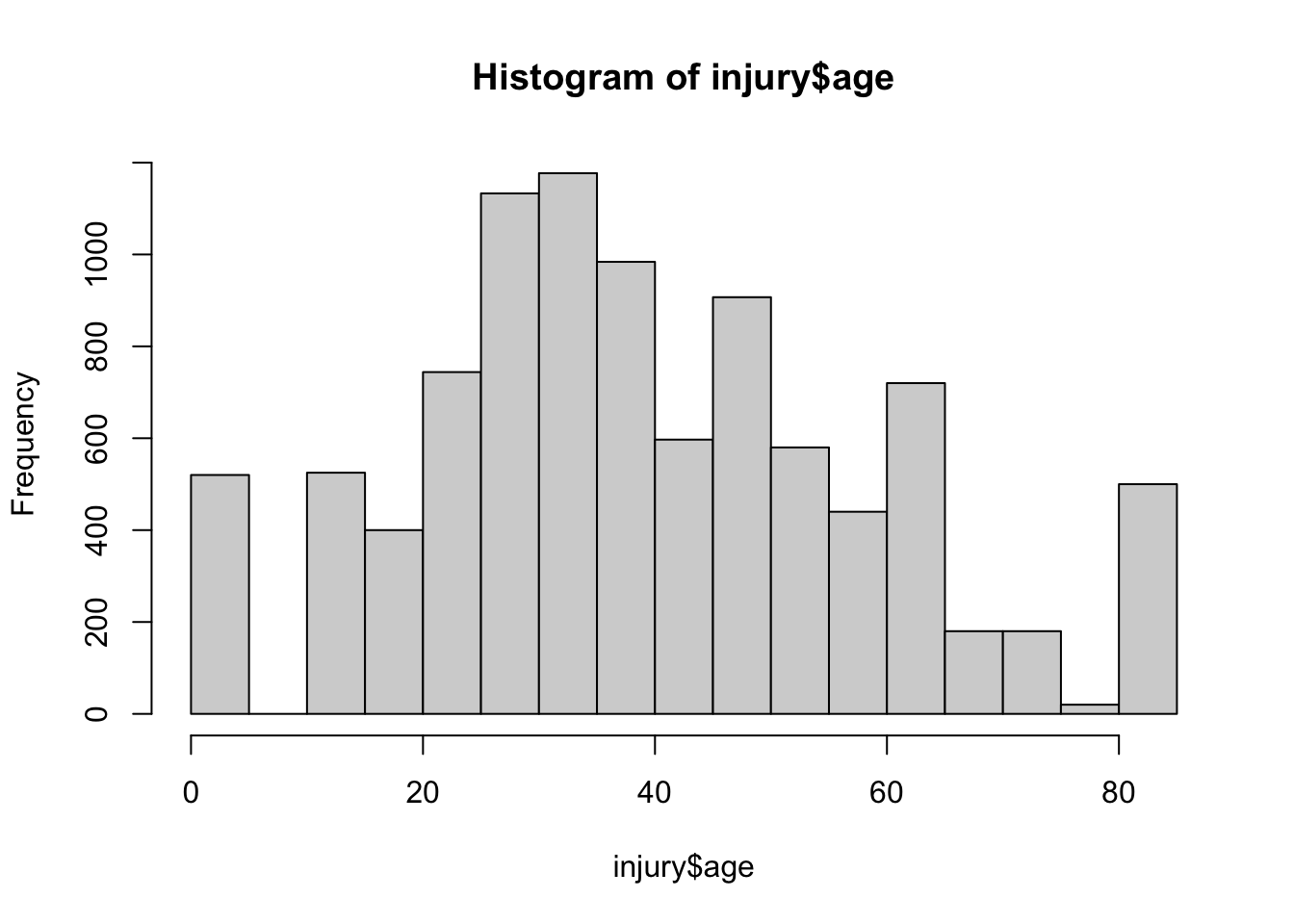
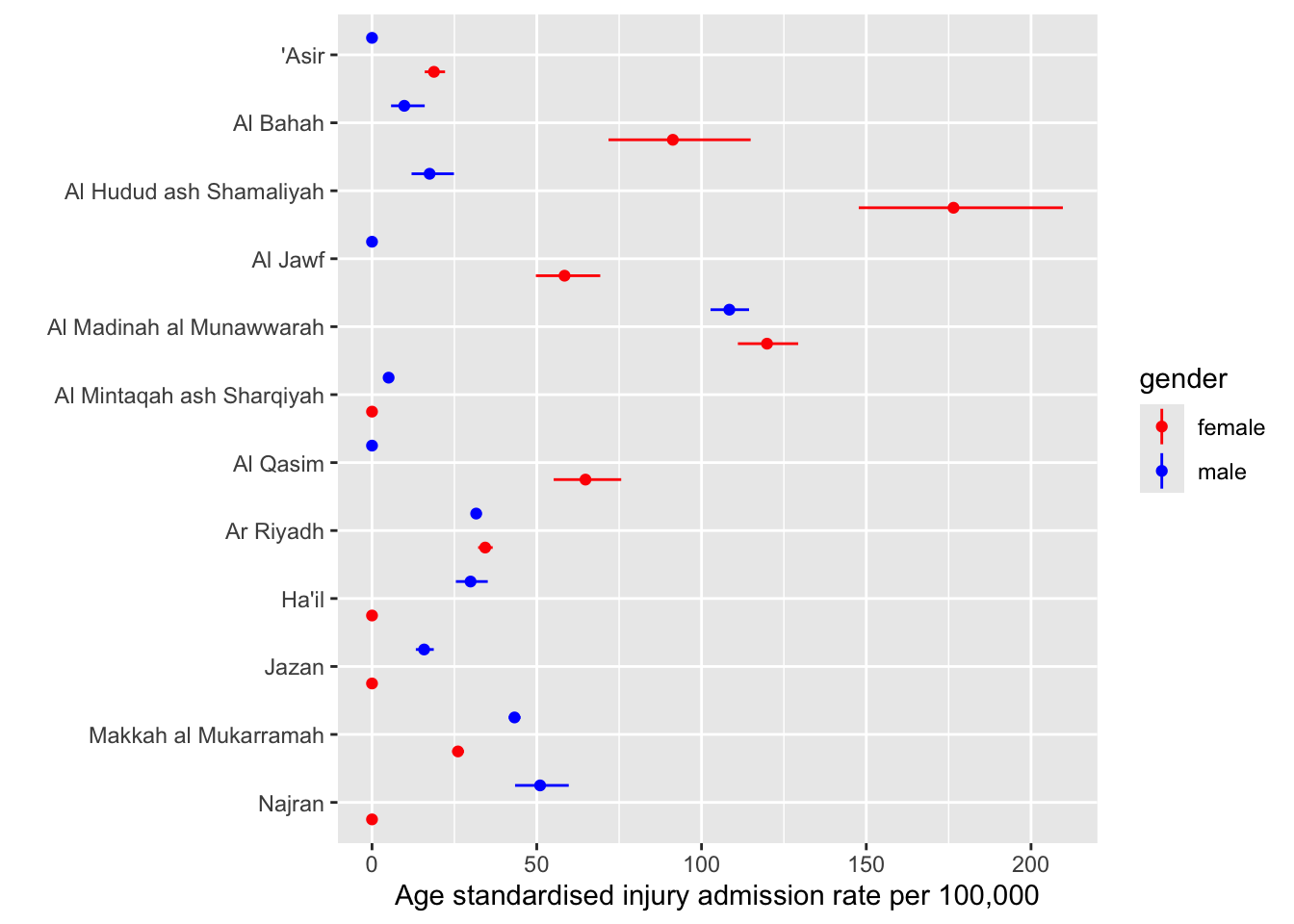
--- title: "Injury" format: html: toc: true toc-location: right editor: visual execute: echo: false output: true warning: false message: false --- ## Injury ```{r} #| label: setup #| include: true needs (data.table, tidyverse, readxl, conflicted)conflicts_prefer (base:: intersect, dplyr:: filter)<- read_rds ("data/counts.rds" )<- read_rds ("data/df.rds" )<- read_csv ("~/proof-of-concept/data/pops_1.csv" )<- df_raw$ pop |> rename (region = 8 ) <- df_raw$ injury ## convert date of birth to age and recode region names ## ``` ```{r} #| label: fig-agehist #| fig-cap: "Age distribution of injury admissions" options (digits = 2 )summary (injury)hist (injury$ age)``` `r nrow(injury)` records, of which `r sum(is.na(injury$age))` (`r 100 * mean(is.na(injury$age))` % ) have missing dates of of birth. These are excluded from analyis```{r} #| label: remove missing ages <- injury |> drop_na (age)``` ```{r} #| label: recode region names to enable linkage to population data <- unique (injury$ Region)``` `r length(unique(injury$Region))` of the 13 regions.```{r} #| label: create age bands <- injury |> mutate (age_band = cut (age, seq (0 , 120 , 5 ), right = FALSE )) ``` ## Aggregate injury data ```{r} #| label: aggregate injury data |> count (Region, diagnosis, code, age_band, Gender) |> pivot_wider (names_from = Gender, values_from = n) |> arrange (code, diagnosis) |> head () |> :: flextable ()``` ## Recode region names to match census names ```{r} #| label: recode regions <- pops |> rename (region = 8 ) |> pluck ("region" ) |> unique ()## do names overlap intersect (census_names, inj_names) ## only 3 regions have names in common ## recode <- injury_dob |> mutate (region = recode (Region, "Asir" = "'Asir" , "Al Baha" = "Al Bahah" ,"Sharqiya" = "Al Mintaqah ash Sharqiyah" ,"Riyadh" = "Ar Riyadh" ,"Al Qassim" = "Al Qasim" ,"Hail" = "Ha'il" , "Makkah" = "Makkah al Mukarramah" ,"Madinah" = "Al Madinah al Munawwarah" ,"Northern Frontier" = "Al Hudud ash Shamaliyah" ))``` ```{r} #| label: recode age bands ## recode age bands ## $ ` Five-Year Age Group ` |> unique ()$ age_band |> unique ()<- injury_dob |> mutate (age_band = recode (age_band, "[0,5)" = "0-4" , "[5,10)" = "5-9" , "[10,15)" = "10-14" , "[15,20)" = "15-19" , "[20,25)" = "20-24" , "[25,30)" = "25-29" , "[30,35)" = "30-34" , "[35,40)" = "35-39" ,"[40,45)" = "40-44" , "[45,50)" = "45-49" ,"[50,55)" = "50-54" , "[55,60)" = "55-59" ,"[60,65)" = "60-64" , "[65,70)" = "65-69" ,"[70,75)" = "70-74" ,"[75,80)" = "75-79" ,"[80,85)" = "80+" ,"[85,90)" = "80+" , "[90,95)" = "80+" , "[95,100)" = "80+" ,"[100,105)" = "80+" , "[105,110)" = "80+" ,"[110,115)" = "80+" , "[115,120)" = "80+" )) ``` ```{r} #| label: aggregate pop data <- pops |> group_by (region, Gender, age_band = ` Five-Year Age Group ` ) |> reframe (tot_pop = sum (Population))``` ```{r} #| label: calculate age-standardised hospitalisation rates - link population data <- injury_dob |> select (region, Gender, age_band, code, diagnosis) |> mutate (age_band = fct_relevel (age_band, "5-9" , after = 1 ))## count by region, ageband, gender and complete missing levels. Fill missing levels with 0 <- injury_dob |> group_by (region, age_band, Gender) |> reframe (n = n ()) |> arrange (region) |> complete (region, age_band, Gender, fill = list (0 )) |> # pivot_wider(names_from = Gender, values_from = n) |> mutate_if (is.numeric, \(x) ifelse (is.na (x), 0 , x))## link to pop data ## <- injury_rate_agg |> full_join (by = c ("Gender" , "region" , "age_band" )) |> drop_na () |> mutate (age_band = fct_relevel (age_band, "5-9" , after = 1 ))$ age_band |> unique ()``` ```{r} #| label: add uncertainty intervals #| eval: false <- injury_rate |> phe_rate (x = n, n = tot_pop)|> arrange (age_band) |> ggplot (aes (age_band, value, group = region, fill = region), ) + geom_col (position = position_dodge ()) + facet_wrap (~ Gender)``` ## Calculate directly age standardised rates for injury admission by region and gender `epitools::ageadjust.direct` .`nest_by` function to group data by region and apply standardiastion function.```{r} #| label: fig-dsr #| fig-cap: "Age standardised injury admission rate per 100,000 by region and gender" needs (epitools)<- pop_agg |> group_by (age_band, Gender) |> reframe (ksa_pop = sum (tot_pop))## Calculate standard population male and female <- ref_pops |> filter (Gender == "Female" ) |> select (ksa_pop)<- ref_pops |> filter (Gender == "Male" ) |> select (ksa_pop)<- injury_rate |> filter (Gender == "Female" ) <- injury_rate |> filter (Gender == "Male" )## add reference population <- irf |> bind_cols (ref = rep (fstd$ ksa_pop, times = 12 )) <- irm |> bind_cols (ref = rep (mstd$ ksa_pop, times = 12 )) ## calculate dsr <- irf |> nest_by (region) |> mutate (dsr = list (epitools:: ageadjust.direct (count = data$ n, pop = data$ tot_pop, stdpop = data$ ref))) |> unnest_wider (dsr)<- irm |> nest_by (region) |> mutate (dsr = list (epitools:: ageadjust.direct (count = data$ n, pop = data$ tot_pop, stdpop = data$ ref))) |> unnest_wider (dsr)<- female_dsr |> mutate (gender = "female" ) |> bind_rows (male_dsr |> mutate (gender = "male" )) |> select (- data)|> write_csv ("data/injury_dsr.csv" )## bind m and f values |> mutate (gender = "female" ) |> bind_rows (male_dsr |> mutate (gender = "male" )) |> ggplot () + geom_point (aes (fct_rev (region), 100000 * adj.rate, colour = gender), position = position_dodge (width = 1 )) + geom_linerange (aes (fct_rev (region), ymin = 100000 * lci, ymax = 100000 * uci, colour = gender), position = position_dodge (width = 1 )) + coord_flip () + labs (y = "Age standardised injury admission rate per 100,000" , x = "" ) + scale_colour_manual (values = c ("red" , "blue" ))```